5 Scopus
Since the amount of documents returned was the same for Scopus and WoS, 358 publications, I chose to work with Scopus. Scopus allows greater control of author addresses via the API rscopus.
Query
358 document results
( TITLE-ABS-KEY ( pirarucu ) OR TITLE-ABS-KEY ( "arapaima gigas" ) OR TITLE-ABS-KEY ( paiche ) ) Search performed on 2022-11-12.
M <- bibliometrix::convert2df(file = 'bibs/scopus_2022-11-12.bib', dbsource = "scopus", format = "bibtex")
rio::export(M, 'rawfiles/m_scopus.rds')
M |>
tibble::as_tibble() |>
dplyr::select(SR_FULL, TI, DI, url) |>
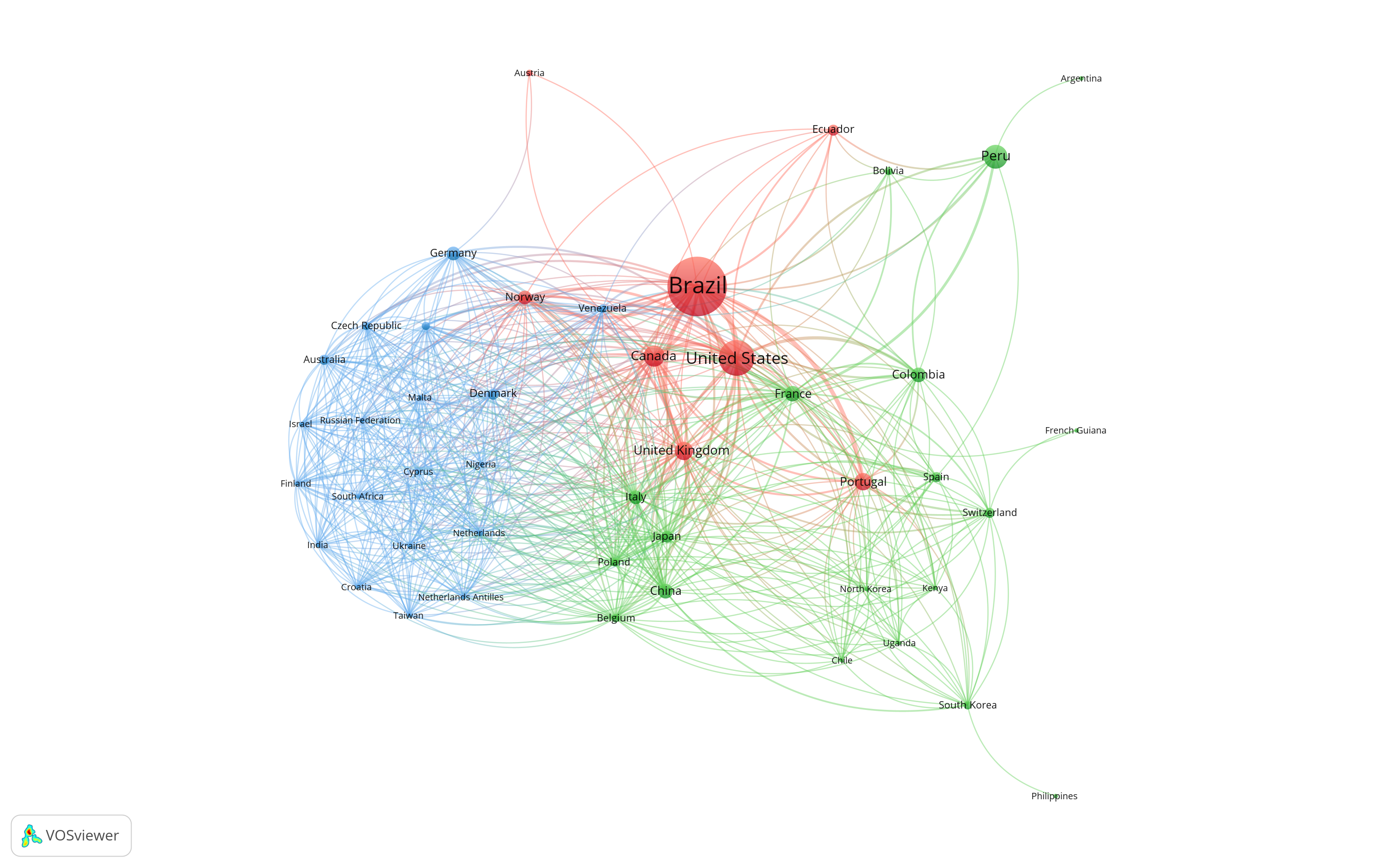
{\(x) rio::export(x, 'rawfiles/data_supplementary.xlsx') }() 5.1 Countries collaboration netowrk
rio::import('rawfiles/m_scopus.rds') -> M
# Address Scopus API
# existe API
rscopus::have_api_key()
# ----------
##
# M[10, ] |> dplyr::pull(TI)
# Query <- "TITLE ('ASSESSMENT OF SMALLHOLDER FARMERS ADAPTIVE CAPACITY TO CLIMATE CHANGE : USE OF A MIXED WEIGHTING SCHEME') AND (LIMIT-TO ( DOCTYPE , 'ar') OR LIMIT-TO ( DOCTYPE , 're' ))"
# rscopus::scopus_search(query = Query,
# view = "COMPLETE",
# count = 200) ->
# x
# rscopusAffiliation(x)
# rscopusAutInsArt(x)
# rscopusAuthors(x)
# ----------
p1 <- "( TITLE ( '"
p2 <- "' ) OR DOI ("
p3 <- ") )"
M |>
dplyr::select(TI, DI) |>
dplyr::mutate(query = paste0(p1, TI, p2, DI, p3)) |>
dplyr::pull(query) ->
query
my_scopus_search <- function(x) {
rscopus::scopus_search(query = x, view = "COMPLETE", count = 200)
}
query |>
purrr::map(purrr::safely(my_scopus_search)) |>
purrr::map(purrr::pluck, 'result') ->
res_previo
res_previo |>
purrr::map(purrr::safely(rscopusAffiliation)) |>
purrr::map(purrr::pluck, 'result') |>
purrr::map(tibble::as_tibble) ->
afiliacao
rio::export(afiliacao, 'rawfiles/m_scopus_afilicao.rds')
query |>
purrr::map(purrr::safely(my_scopus_search)) |>
purrr::map(purrr::pluck, 'result') ->
res
rio::export(res, 'rawfiles/res.rds')
res |>
purrr::map(purrr::safely(rscopusSponsor)) |>
purrr::map(purrr::pluck, 'result') |>
dplyr::bind_rows() |>
dplyr::group_by(fund_sponsor) |>
dplyr::tally(name = 'qtde', sort = T) |>
{\(x) rio::export(x, 'rawfiles/scopus_financiadores.xlsx') }() rio::import('rawfiles/m_scopus.rds') ->
M
rio::import('rawfiles/m_scopus_afilicao.rds') ->
afiliacao
M$id <- as.character(1:nrow(M))
# ------------
#
afiliacao |>
purrr::map(janitor::clean_names) |>
dplyr::bind_rows(.id = 'id') |>
dplyr::group_by(id) |>
tidyr::nest() |>
dplyr::rename(affiliation = data) |>
dplyr::ungroup() |>
dplyr::right_join(M) ->
M2
M2 |>
dplyr::select(SR, id, affiliation) |>
tidyr::unnest_wider(affiliation) |>
tidyr::unnest_longer(affiliation_country) |>
dplyr::select(SR, id, affiliation_country) |>
dplyr::distinct(.keep_all = TRUE) |>
dplyr::group_by(SR, id) |>
dplyr::summarise(qtde_paises = dplyr::n()) |>
dplyr::ungroup() ->
artigo_qtde_paises
afiliacao |>
purrr::compact() |>
purrr::map(~ .x |> janitor::clean_names()) |>
purrr::map(~ .x |> dplyr::pull(affiliation_country)) |>
purrr::map(function(x) {expand.grid.unique(x, x, include.equals = F)}) |>
dplyr::bind_rows() |>
tibble::as_tibble() ->
ide
c(ide$V1, ide$V2) |>
unique() |>
sort() ->
idv
ide |>
dplyr::filter(!is.na(V1)) |>
dplyr::filter(!is.na(V2)) ->
ide2
igraph::graph.data.frame(ide2, directed = FALSE, vertices = idv) |>
{\(x) igraph::graph.adjacency(igraph::get.adjacency(x), weighted = TRUE)}() |>
{\(x) igraph::simplify(x, remove.multiple = T, edge.attr.comb = list(weight = 'sum'))}() |>
tidygraph::as_tbl_graph() ->
net
M2 |>
dplyr::select(SR, id, affiliation) |>
tidyr::unnest_wider(affiliation) |>
tidyr::unnest_longer(affiliation_country) |>
dplyr::select(SR, id, affiliation_country) |>
dplyr::distinct(.keep_all = TRUE) |>
dplyr::group_by(affiliation_country) |>
dplyr::summarise(qtde_artigos = n()) |>
dplyr::rename(name = affiliation_country) |>
dplyr::ungroup() ->
paises_artigos
net |>
tidygraph::activate(nodes) |>
dplyr::left_join(paises_artigos) |>
dplyr::arrange(desc(qtde_artigos)) ->
net2
net2 |>
tidygraph::activate(nodes) |>
dplyr::mutate(degree = degree(net2),
closeness = closeness(net2),
betweenness = betweenness(net2)) |>
tibble::as_tibble() ->
net2_atributos
# ---------
## sjr
readr::read_csv2('rawfiles/scimagojr_2019_filtrado.csv') |>
janitor::clean_names() |>
dplyr::arrange(desc(sjr)) |>
tidyr::separate_rows(issn, sep = ',') |>
dplyr::mutate(issn = stringr::str_trim(issn)) |>
dplyr::distinct(issn, .keep_all = TRUE) ->
sjr
M2 |>
dplyr::select(SR, SN, id, affiliation) |>
tidyr::unnest_wider(affiliation) |>
tidyr::unnest_longer(affiliation_country) |>
dplyr::select(SN, id, affiliation_country) |>
dplyr::rename(issn = SN) |>
dplyr::distinct(.keep_all = TRUE) |>
left_join(sjr) |>
dplyr::mutate(sjr = ifelse(is.na(sjr), 20, sjr)) ->
m2_sjr
m2_sjr |>
dplyr::select(id, affiliation_country, sjr) |>
dplyr::filter(!is.na(affiliation_country)) |>
dplyr::group_by(id, affiliation_country) |>
dplyr::slice_head(n = 1) |>
dplyr::ungroup() |>
dplyr::group_by(affiliation_country) |>
dplyr::summarise(sjr = sum(sjr)) |>
dplyr::rename(name = affiliation_country) ->
sjr_scopus
net2_atributos |>
dplyr::full_join(sjr_scopus) |>
dplyr::mutate(sjr_medio = sjr / qtde_artigos) |>
dplyr::relocate(name, qtde_artigos, sjr, sjr_medio, degree, closeness, betweenness) ->
net2_atributos2
rio::export(net2_atributos2, 'rawfiles/m_scopus_col_cou_atributos2.xlsx')
artigos3 |>
dplyr::left_join(sjr %>>% dplyr::select(issn, sjr)) |>
dplyr::mutate(sjr = ifelse(is.na(sjr), 20, sjr)) ->
artigos4
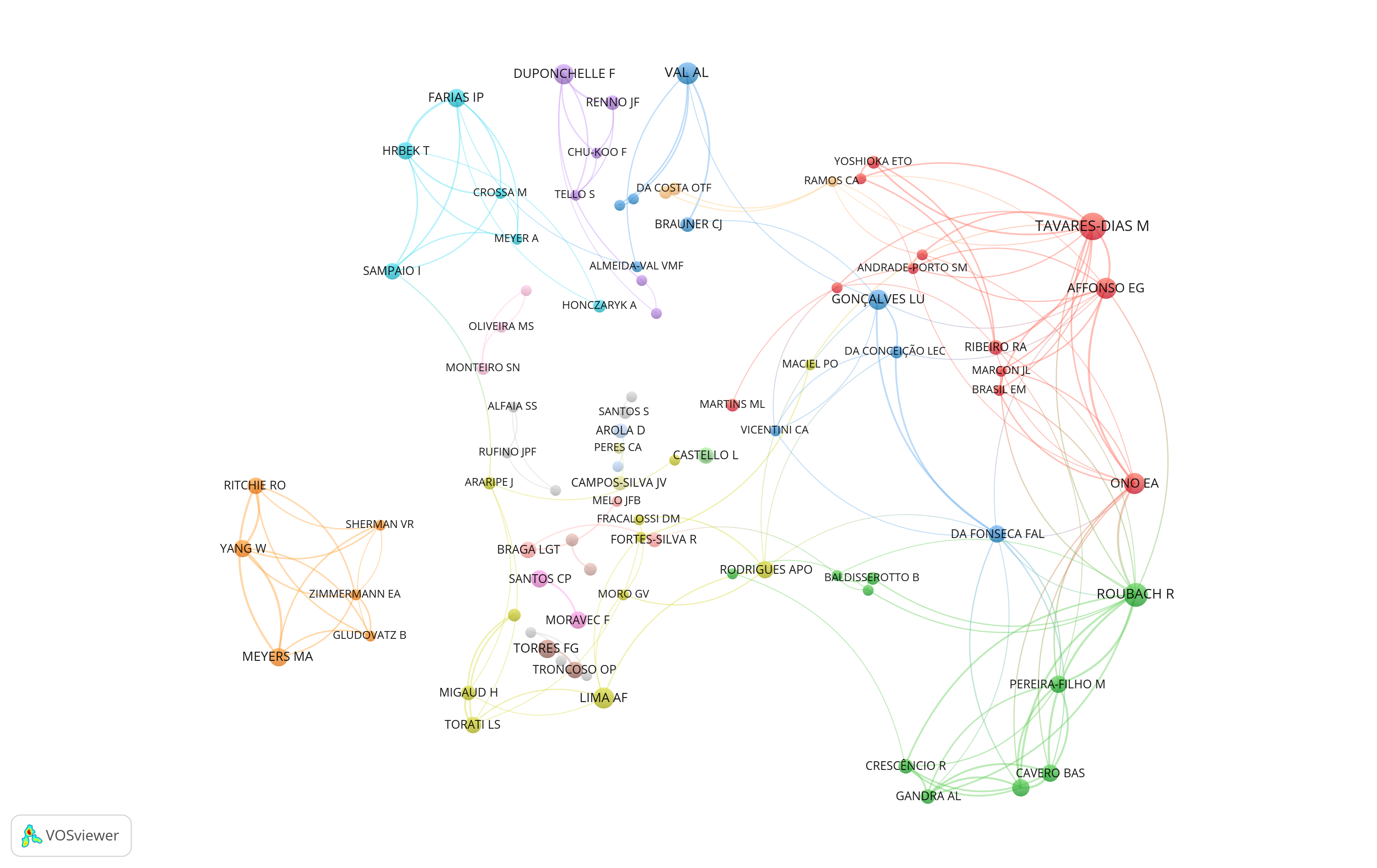
eb <- igraph::cluster_louvain(as.undirected(net2))
net2 |>
tidygraph::activate(nodes) |>
dplyr::mutate(group = eb$membership) ->
net3
net3 |>
tidygraph::activate(nodes) |>
group_by(eb) |>
slice_head(n = 4) |>
print(n = Inf)## Error in (function (cond) : error in evaluating the argument 'x' in selecting a method for function 'print': Must group by variables found in `.data`.
## ✖ Column `eb` is not found.5.2 Growth rate
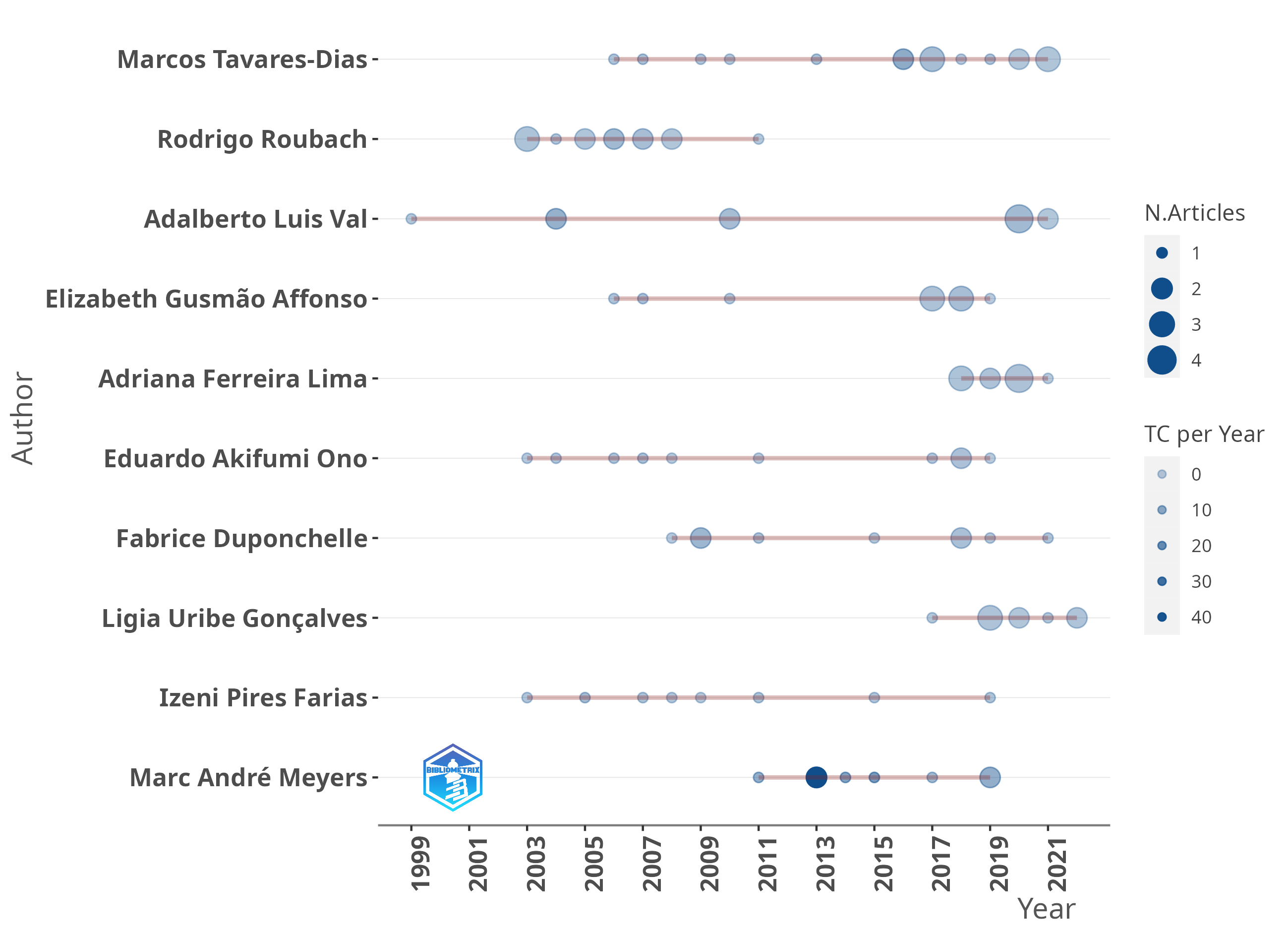
Growth rate for documents obtained from Scopus for the period 2000 to 2021.
M <- rio::import('rawfiles/m_scopus.rds')
M %>>%
count(PY, sort = F, name = 'Papers') %>>%
(~ d0) %>>%
dplyr::filter(PY %in% c(2000:2020)) %>>%
dplyr::arrange(PY) %>>%
dplyr::mutate(trend = 1:n()) %>>%
(. -> d)
d$lnp <- log(d$Papers)
m1 <- lm(lnp ~ trend, data = d)
# summary(m1)
beta0 <- m1$coefficients[[1]]
beta1 <- m1$coefficients[[2]]
# 2000
m2 <- nls(Papers ~ b0 * exp(b1 * (PY - 2000)), start = list(b0 = beta0, b1 = beta1), data = d)
# summary(m2)
d$predicted <- 1.77285 * exp(0.15751 * (d$PY - 2000))
# (exp(0.15751) - 1) * 100
# log(2) / 0.15751
d %>>%
mutate(Year = PY) %>>%
mutate(predicted = round(predicted, 0)) %>>%
(. -> d)
periodo <- 2000:2024
predicted <- tibble::tibble(PY = periodo, Predicted = round(1.77285 * exp(0.15751 * (periodo - 2000)), 0))
dplyr::full_join(d0, predicted) |>
dplyr::rename(Year = PY) |>
dplyr::filter(Year %in% 2000:2020) ->
d2
# p <- ggplot2::ggplot(d2, aes(x = Year, y = Papers)) +
# ggplot2::geom_line(aes(x = Year, y = Papers, colour = "Papers Scopus")) +
# ggplot2::geom_point(aes(y = Papers, color = 'Papers Scopus')) +
# ggplot2::geom_line(aes(y = Predicted, color = 'Predicted Scopus'), linetype = 'longdash') +
# ggplot2::geom_point(aes(y = Predicted, color = 'Predicted Scopus')) +
# scale_x_continuous(name = 'Year', breaks = seq(2000, 2020, by = 2), limits = c(2000, 2020)) +
# scale_y_continuous(name = 'Papers', breaks = seq(0, 45, by = 5), limits = c(0, 45)) +
# scale_color_manual(name = "Publications", values = c("Predicted Scopus" = "red", "Papers Scopus" = "black")) +
# theme_bw() +
# ylab('Papers') +
# theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1), legend.position = "bottom")
# p + ggplot2::annotate("text", x = 2009, y = 40, size = 3, label = 'Predicted Y_p = 1.77 * e ^ 0.15 (year-2000)', parse = F)
# ggsave('images/taxa-crescimento-scopus.png', width = 10, height = 10, units = c("cm"))
highcharter::hchart(d2, "column", hcaes(x = Year, y = Papers), name = "Publications", showInLegend = TRUE) %>>%
highcharter::hc_add_series(d2, "line", hcaes(x = Year, y = Predicted), name = "Predicted", showInLegend = TRUE) %>>%
highcharter::hc_add_theme(hc_theme_google()) %>>%
highcharter::hc_navigator(enabled = TRUE) %>>%
highcharter::hc_exporting(enabled = TRUE, filename = 'groups_growth') %>>%
highcharter::hc_xAxis(plotBands = list(list(from = 2021, to = 2021, color = "#330000")))- Growth Rate 17.05%
- Doubling time 4.4 Years
Growth rate of the entire Scopus.
d0 <- read.table('rawfiles/Scopus-38425968-Analyze-Year.csv', header = T, sep = ',', skip = 3)
d0 |>
dplyr::mutate(Papers = Papers / 100000) |>
dplyr::filter(PY %in% c(2000:2021)) %>>%
dplyr::arrange(PY) %>>%
dplyr::mutate(trend = 1:n()) %>>%
(. -> d)
d$lnp <- log(d$Papers)
m1 <- lm(lnp ~ trend, data = d)
# summary(m1)
beta0 <- m1$coefficients[[1]]
beta1 <- m1$coefficients[[2]]
# 2000
m2 <- nls(Papers ~ b0 * exp(b1 * (PY - 2000)), start = list(b0 = beta0, b1 = beta1), data = d)
# summary(m2)
d$predicted <- 1.26206 * exp(0.169719 * (d$PY - 1990))
# (exp(0.052582) - 1) * 100
# log(2) / 0.052582- Growth Rate 5.5%
- Doubling time 13 Years
5.3 Top Journals
results <- bibliometrix::biblioAnalysis(M, sep = ";")
S <- summary(object = results, k = 10, pause = F, verbose = F)
S$MostRelSources |>
datatable(
extensions = 'Buttons',
rownames = F,
options = list(
dom = 'Bfrtip',
pageLength = 10,
buttons = list(list(
extend = 'collection',
buttons = list(list(extend = 'csv', filename = 'data'),
list(extend = 'excel', filename = 'data')),
text = 'Download')))) 5.4 Most cited papers
M |>
tibble::as_tibble() |>
dplyr::select(SR, AU, PY, TI, DI, TC, DE, AB, SO) |>
{\(x) rio::export(x, 'rawfiles/m_scopus_resumo.xlsx') }()
S$MostCitedPapers |>
datatable(
extensions = 'Buttons',
rownames = F,
options = list(
dom = 'Bfrtip',
pageLength = 10,
buttons = list(list(
extend = 'collection',
buttons = list(list(extend = 'csv', filename = 'data'),
list(extend = 'excel', filename = 'data')),
text = 'Download')))) 5.6 Articles by country
Number of articles with at least one author from the country.
paises_artigos |>
dplyr::filter(!is.na(name)) |>
dplyr::arrange(desc(qtde_artigos)) |>
datatable(
extensions = 'Buttons',
rownames = F,
options = list(
dom = 'Bfrtip',
pageLength = 10,
buttons = list(list(
extend = 'collection',
buttons = list(list(extend = 'csv', filename = 'data'),
list(extend = 'excel', filename = 'data')),
text = 'Download'))))