Overview
birddog helps you detect emergence and trace
trajectories in scientific literature and patents. It reads datasets
from OpenAlex and Web of Science (WoS), builds citation-based networks,
identifies groups, and summarizes their dynamics.
A stable release is planned for CRAN. The development version is available on GitHub: https://github.com/roneyfraga/birddog.
Data sources
-
birddogsupports:- OpenAlex: browser search with CSV export, or API via openalexR.
-
Web of
Science: multiple export formats (
.bib,.ris, plain-text.txt, tab-delimited.txt).
OpenAlex via API or CSV
You can paste a URL from openalex.org and prefix it with
https://api. to obtain the API endpoint.
# install.packages("openalexR")
library(openalexR)
# Example: all publications in the Journal of Evolutionary Economics
url_web <- "https://openalex.org/works?page=1&filter=primary_location.source.id:s121026525"
url_api <- "https://api.openalex.org/works?page=1&filter=primary_location.source.id:s121026525"
openalexR::oa_request(query_url = url_api) |>
openalexR::oa2df(entity = "works") |>
birddog::read_openalex(format = "api") ->
file
M <- birddog::read_openalex(file, format = "api")Web of Science (WoS)
WoS allows exporting in several formats. birddog can
read:
# openalex: csv
M <- birddog::read_openalex('http://roneyfraga.com/volume/keep_it/birddog-data/openalex-works-2025-05-28T23-12-11.csv', format = "csv")
# wos: txt-plain-text
M <- birddog::read_wos('http://roneyfraga.com/volume/keep_it/birddog-data/wos-savedrecs-plain-text.txt', format = "txt-plain-text")
# wos: txt-tab-delimited
M <- birddog::read_wos('http://roneyfraga.com/volume/keep_it/birddog-data/wos-savedrecs-tab-delimited.txt', format = "txt-tab-delimited")
# wos: ris
M <- birddog::read_wos('http://roneyfraga.com/volume/keep_it/birddog-data/wos-savedrecs.ris', format = "ris")
# wos: bib
M <- birddog::read_wos('http://roneyfraga.com/volume/keep_it/birddog-data/wos-savedrecs.bib', format = "bib", normalized_names = TRUE)Example dataset
To save processing time, we’ll use a pre-saved WoS sample available in https://roneyfraga.com/volume/keep_it/birddog-data/wos-sugarcane-m.rds.
12,689 results from Web of Science Core Collection for:
"sugarcane" AND ("straw" OR "bagasse" OR "filter cake" OR "press mud" OR "pressmud cake" OR "molasses" OR "vinasse" OR "dried yeast" OR "fusel oil")Download with the query above in 2023-09-27. Full query here: https://www.webofscience.com/wos/woscc/summary/0fa06733-b4aa-4348-854d-a799cdad2c68-a711a88c/relevance/1.
# bibs <- fs::dir_ls('~/Sync/birddog-data/bibs-sugarcane/', glob = '*.bib$')
#
# tictoc::tic()
# bibs |>
# purrr::map(\(x) birddog::read_wos(x, format = "bib")) |>
# dplyr::bind_rows() |>
# dplyr::distinct(DI2, .keep_all = T) ->
# M
# tictoc::toc()
# 62 sec
url_m <- 'https://roneyfraga.com/volume/keep_it/birddog-data/wos-sugarcane-m.rds'
M <- readRDS(url(url_m))
dplyr::glimpse(M)
#> Rows: 11,512
#> Columns: 50
#> $ AU <chr> "Hernandez-Perez, Andres Felipe and de Arru…
#> $ TI <chr> "Sugarcane straw as a feedstock for xylitol…
#> $ SO <chr> "BRAZILIAN JOURNAL OF MICROBIOLOGY", NA, "B…
#> $ PY <dbl> 2016, 2016, 2013, 2008, 2012, 2022, 2020, 2…
#> $ AB <chr> "Sugarcane straw has become an available li…
#> $ DT <chr> "Article", "Proceedings Paper", "Article", …
#> $ DI <chr> "10.1016/j.bjm.2016.01.019", "10.1016/j.pro…
#> $ DI2 <chr> "101016JBJM201601019", "101016JPROENG201606…
#> $ DE <chr> "Sugarcane straw; Hemicellulosic hydrolyzat…
#> $ ID <chr> "BAGASSE HYDROLYSATE; ACETIC-ACID; FERMENTA…
#> $ SC <chr> "Microbiology", "Engineering; Materials Sci…
#> $ CR <chr> "Anonymous], 2019, COMP NAC AB AC SAFR; Arr…
#> $ TC <chr> "51", "75", "98", "74", "0", "2", "39", "2"…
#> $ JI <chr> "Braz. J. Microbiol.", NA, "Bioresour. Tech…
#> $ SR <chr> "WOS:000376016600030", "WOS:000387712600117…
#> $ DB <chr> "wos_bib_normalized_normalized_names", "wos…
#> $ volume <chr> "47", "148", "131", "148", NA, "57", "25", …
#> $ number <chr> "2", NA, NA, "1-3", NA, "2, SI", "3", "5", …
#> $ pages <chr> "489-496", "839-846", "357-364", "45-58", "…
#> $ month <chr> "APR-JUN", NA, "MAR", "MAR", NA, "FEB", "FE…
#> $ publisher <chr> "SPRINGER", "ELSEVIER SCIENCE BV", "ELSEVIE…
#> $ address <chr> "233 SPRING ST, NEW YORK, NY 10013 USA", "S…
#> $ language <chr> "English", "English", "English", "English",…
#> $ C1 <chr> "Hernández-Pérez, AF (Corresponding Author)…
#> $ issn <chr> "1517-8382", "1877-7058", "0960-8524", "027…
#> $ eissn <chr> "1678-4405", NA, "1873-2976", "1559-0291", …
#> $ web_of_science_categories <chr> "Microbiology", "Engineering, Industrial; M…
#> $ author_email <chr> "[email protected]", "[email protected]…
#> $ affiliations <chr> "Universidade de Sao Paulo", "Universiti Te…
#> $ researcher_id_numbers <chr> "Pérez, Andrés Felipe Hernández/AAN-5546-20…
#> $ orcid_numbers <chr> "Pérez, Andrés Felipe Hernández/0000-0002-5…
#> $ funding_acknowledgement <chr> "FAPESP (Fundacao do amparo a pesquisa do e…
#> $ funding_text <chr> "This work was financially supported by the…
#> $ number_of_cited_references <chr> "39", "12", "36", "35", "13", "36", "49", "…
#> $ usage_count_last_180_days <chr> "0", "2", "1", "1", "0", "5", "0", "3", "1"…
#> $ usage_count_since_2013 <chr> "12", "7", "133", "40", "4", "34", "19", "9…
#> $ doc_delivery_number <chr> "DM0EV", "BG2UR", "118KK", "289MD", "BGL38"…
#> $ web_of_science_index <chr> "Science Citation Index Expanded (SCI-EXPAN…
#> $ oa <chr> "hybrid, Green Published", "gold", NA, NA, …
#> $ da <chr> "2023-11-14", "2023-11-14", "2023-11-14", "…
#> $ editor <chr> NA, "Bustam, MA and Man, Z and Keong, LK an…
#> $ booktitle <chr> NA, "PROCEEDING OF 4TH INTERNATIONAL CONFER…
#> $ series <chr> NA, "Procedia Engineering", NA, NA, NA, NA,…
#> $ note <chr> NA, "4th International Conference on Proces…
#> $ isbn <chr> NA, NA, NA, NA, "978-7-5019-9043-6", NA, NA…
#> $ early_access_date <chr> NA, NA, NA, NA, NA, "DEC 2021", NA, NA, "AU…
#> $ article_number <chr> NA, NA, NA, NA, NA, NA, "623", "PII S174217…
#> $ book_group_author <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ book_author <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ meeting <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…Build a citation network
You can build either a direct citation network or use bibliographic coupling.
Direct citation highlights time-ordered influence; bibliographic coupling captures proximity in topics via shared references.
# Direct citation
# net <- birddog::sniff_network(M, type = "direct citation")
# Bibliographic coupling
net <- birddog::sniff_network(M, type = "bibliographic coupling")
net |>
tidygraph::activate(nodes) |>
dplyr::select(name, AU, PY, TI, TC) |>
dplyr::arrange(dplyr::desc(TC))
#> # A tbl_graph: 11416 nodes and 2659060 edges
#> #
#> # An undirected simple graph with 115 components
#> #
#> # Node Data: 11,416 × 5 (active)
#> name AU PY TI TC
#> <chr> <chr> <dbl> <chr> <chr>
#> 1 101016JAPENERGY201809135 Chen, Wei-Hsin and Lin, Bo-Jhih… 2018 Hygr… 99
#> 2 101016JBEJ200602009 Rahman, S. H. A. and Choudhury,… 2006 Prod… 99
#> 3 101016JBIOMBIOE201606017 Zhu, Zongyuan and Rezende, Cami… 2016 Effi… 99
#> 4 101016JCARBPOL201407052 Szczerbowski, Danielle and Pita… 2014 Suga… 99
#> 5 101016JCARBPOL201607071 Candido, R. G. and Goncalves, A… 2016 Synt… 99
#> 6 101016JCARBPOL201808081 Harini, K. and Ramya, K. and Su… 2018 Extr… 99
#> 7 101016JPBIOMOLBIO201807011 Meili, L. and Lins, P. V. S. an… 2019 Adso… 99
#> 8 101016JRSER201405036 Rocha, Mateus Henrique and Capa… 2014 Life… 99
#> 9 101016S0032959200001503 Patil, YB and Paknikar, KM 2000 Deve… 99
#> 10 101021IE401286Z Subhedar, Preeti B. and Gogate,… 2013 Inte… 99
#> # ℹ 11,406 more rows
#> #
#> # Edge Data: 2,659,060 × 3
#> from to weight
#> <int> <int> <dbl>
#> 1 2387 6371 1
#> 2 588 2387 1
#> 3 2387 5633 1
#> # ℹ 2,659,057 more rowsComponents
The analysis of components is important to eliminate disconnected documents that do not share the same bibliographic references. However, if more than one component with a high number of documents exists, it may indicate the presence of two disconnected scientific literatures.
comps <- birddog::sniff_components(net)
names(comps)
#> [1] "components" "network"
comps$components |>
dplyr::slice_head(n = 5) |>
gt::gt()| component | quantity_publications | average_age |
|---|---|---|
| component1 | 11298 | 2017.469 |
| component2 | 2 | 2012.500 |
| component3 | 2 | 1993.500 |
| component4 | 2 | 1997.000 |
| component5 | 2 | 2020.000 |
Groups (community detection)
birddog::sniff_groups(
comps,
algorithm = 'fast_greedy',
min_group_size = 30,
groups_short_name = TRUE) ->
groups
names(groups)
#> [1] "aggregate" "network" "pubs_by_year"
groups$aggregate |>
gt::gt()| group | quantity_papers | average_age |
|---|---|---|
| g01 | 3022 | 2017.690 |
| g02 | 2861 | 2017.528 |
| g03 | 1966 | 2018.080 |
| g04 | 1819 | 2016.885 |
| g05 | 968 | 2019.461 |
| g06 | 414 | 2014.587 |
| g07 | 204 | 2009.446 |
Group attributes
It helps to understand the structure of the groups.
birddog::sniff_groups_attributes(
groups,
growth_rate_period = 2010:2022,
show_results = FALSE) ->
groups_attributes
names(groups_attributes)
#> [1] "attributes_table" "regression"
groups_attributes$attributes_table| Groups Attributes | |||||
| Group | Publications | Average age1 | Growth rate2 | Doubling time3 | Horizon plot4 |
|---|---|---|---|---|---|
| g01 | 3022 | 2017+8m | 13.7 | 5y+5m | |
| g02 | 2861 | 2017+6m | 15.3 | 5y+11m | |
| g03 | 1966 | 2018+1m | 10.6 | 7y+11m | |
| g04 | 1819 | 2016+11m | 20.1 | 4y+10m | |
| g05 | 968 | 2019+6m | 30.3 | 3y+7m | |
| g06 | 414 | 2014+7m | -0.6 | NAy+NAm | |
| g07 | 204 | 2009+5m | 13.6 | 5y+5m | |
| Source: Web of Science. Data extracted, organized and estimated by the authors. | |||||
| 1 Average publication year: For example, '2016+7m' means that the articles were published, on average, in 2016 plus seven months. | |||||
| 2 Growth rate percentage year. Calculated by exp(b1)-1 where b1 is the econometric model coefficient. Time span, 2010 until 2022. | |||||
| 3 y = years, m = months. Calculated by ln(2)/b1 where b1 is the econometric model coefficient. | |||||
| 4 Publications between 2010 and 2022. Chart type horizon plot. | |||||
Group content: keywords
It contributes to understanding the content of each group.
groups_keywords <- birddog::sniff_groups_keywords(groups)
groups_keywords |>
DT::datatable(
rownames = FALSE,
filter = 'bottom',
extensions = 'Buttons',
escape = FALSE,
options = list(dom = 'Blfrtip', pageLength = 5)
)Group content: NLP
This step can be time-consuming. Consider precomputing and saving results.
# tictoc::tic()
# groups_terms <- sniff_groups_terms(groups, algorithm = 'phrase')
# tictoc::toc()
# 34 min
groups_terms <- readRDS('~/Sync/birddog-data/wos-sugarcane-groups-terms.rds')
names(groups_terms)
#> [1] "terms_by_group" "terms_table"
groups_terms$terms_table |>
DT::datatable(
rownames = FALSE,
filter = 'bottom',
extensions = 'Buttons',
escape = FALSE,
options = list(dom = 'Blfrtip', pageLength = 5)
)Prestige: hubs
The calculation is slow. Be patient.
# tictoc::tic()
# groups_hubs <- sniff_groups_hubs(groups)
# tictoc::toc()
# 19 min
groups_hubs <- readRDS('~/Sync/birddog-data/wos-sugarcane-groups-hubs.rds')
groups_hubs |>
dplyr::filter(zone != 'noHub') |>
dplyr::left_join(groups$network |> tidygraph::activate(nodes) |> tibble::as_tibble() |> dplyr::select(SR, PY), by = 'SR') |>
dplyr::mutate(Zi = round(Zi, digits = 2), Pi = round(Pi, digits = 2)) |>
dplyr::mutate(SR = paste0('<a href="https://www.webofscience.com/wos/alldb/full-record/', SR, '">', SR, '</a>')) |>
DT::datatable(
rownames = FALSE,
filter = 'bottom',
extensions = 'Buttons',
escape = FALSE,
options = list(dom = 'Blfrtip', pageLength = 10)
)Group evolution (trajectories)
# tictoc::tic()
# groups_cumulative <- sniff_groups_cumulative(groups)
# tictoc::toc()
# 2 min
groups_cumulative <- readRDS('~/Sync/birddog-data/wos-sugarcane-groups-cumulative.rds')
suppressMessages({
groups_cumulative_trajectories <- birddog::sniff_groups_trajectories(groups_cumulative)
})
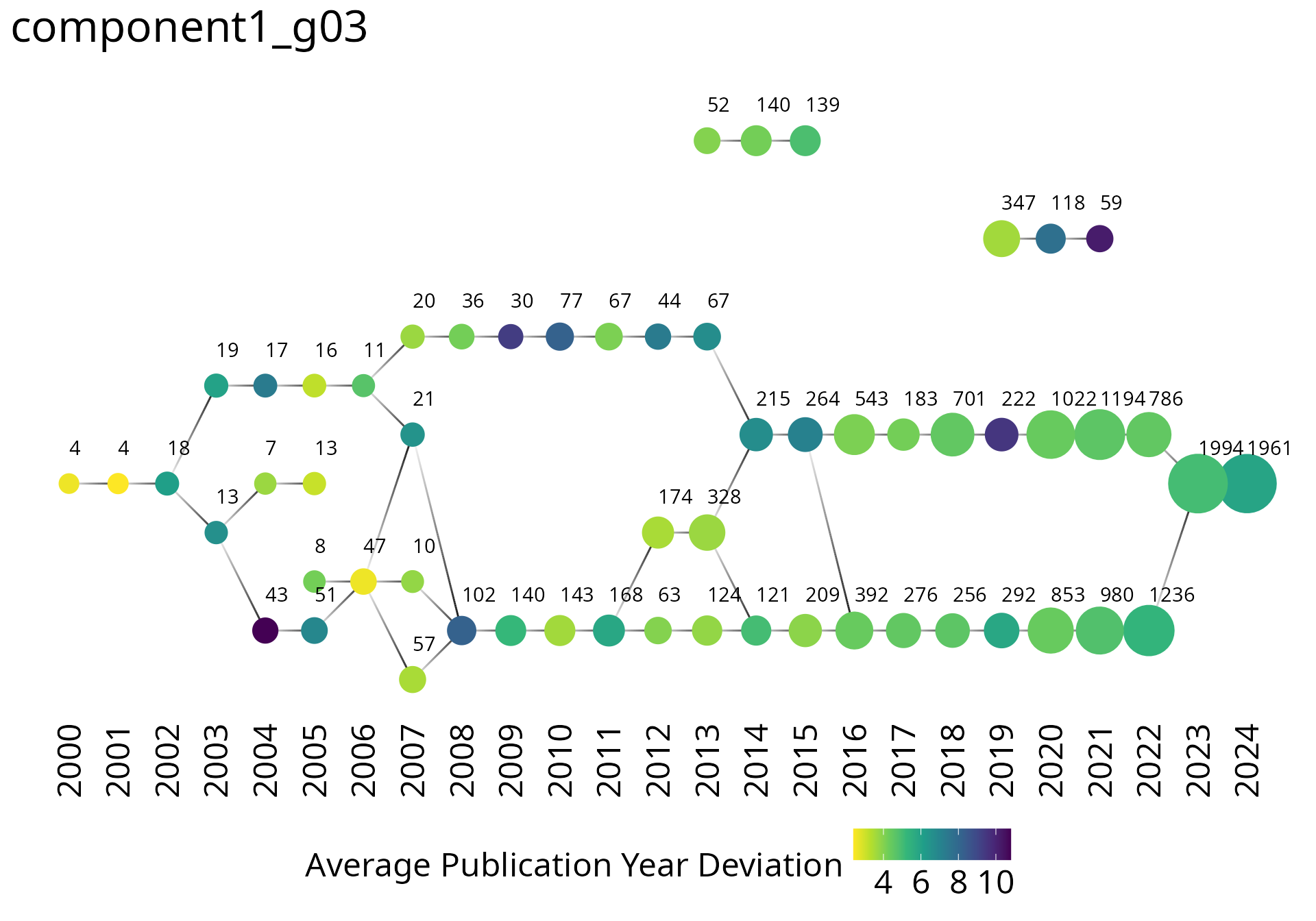
plot_group_trajectories_2d(
groups_cumulative_trajectories,
group = 'component1_g03',
label_vertical_position = -2
)
plot_group_trajectories_3d(
groups_cumulative_trajectories,
group = 'component1_g03'
)Citation growth per document
# tictoc::tic()
# groups_cumulative_citations <- sniff_groups_cumulative_citations(groups, min_citations = 2)
# tictoc::toc()
# 11 min
groups_cumulative_citations <- readRDS('~/Sync/birddog-data/wos-sugarcane-groups-cumulative-citations.rds')
# First, create the data frame
groups_cumulative_citations |>
purrr::map(\(x)
x |>
dplyr::select(- citations_by_year) |>
dplyr::arrange(dplyr::desc(growth_power)) |>
dplyr::slice_head(n = 50)) |>
dplyr::bind_rows() |>
dplyr::mutate(SR = paste0('<a href="https://www.webofscience.com/wos/alldb/full-record/', SR, '">', SR, '</a>')) ->
df
n_cols <- ncol(df)
hidden_cols <- 7:(n_cols - 1) # Hide columns 8 to second-to-last
# Create the datatable
DT::datatable(df, rownames = FALSE, filter = 'bottom', extensions = c('Buttons', 'ColReorder'), escape = FALSE,
options = list(
dom = 'Blfrtip',
pageLength = 10,
columnDefs = list(
list(visible = FALSE, targets = hidden_cols),
list(className = 'dt-center', targets = '_all')
),
buttons = list( list( extend = 'colvis', text = 'Show/Hide Columns', columns = hidden_cols))
))Topic modeling (STM)
Detect topics within a group with Structural Topic Modeling. Here, we create topics (sub-groups) based on linguistic similarities.
# g01
# tictoc::tic()
# groups_stm_prepare_g01 <- sniff_groups_stm_prepare(groups, group_to_stm = 'g01')
# tictoc::toc()
# 21 min
groups_stm_prepare <- readRDS('~/Sync/birddog-data/wos-sugarcane-groups-stm-prepare-g01.rds')
names(groups_stm_prepare)
#> [1] "result" "plots" "data" "parameters"
groups_stm_prepare$plots
#> $metrics_by_k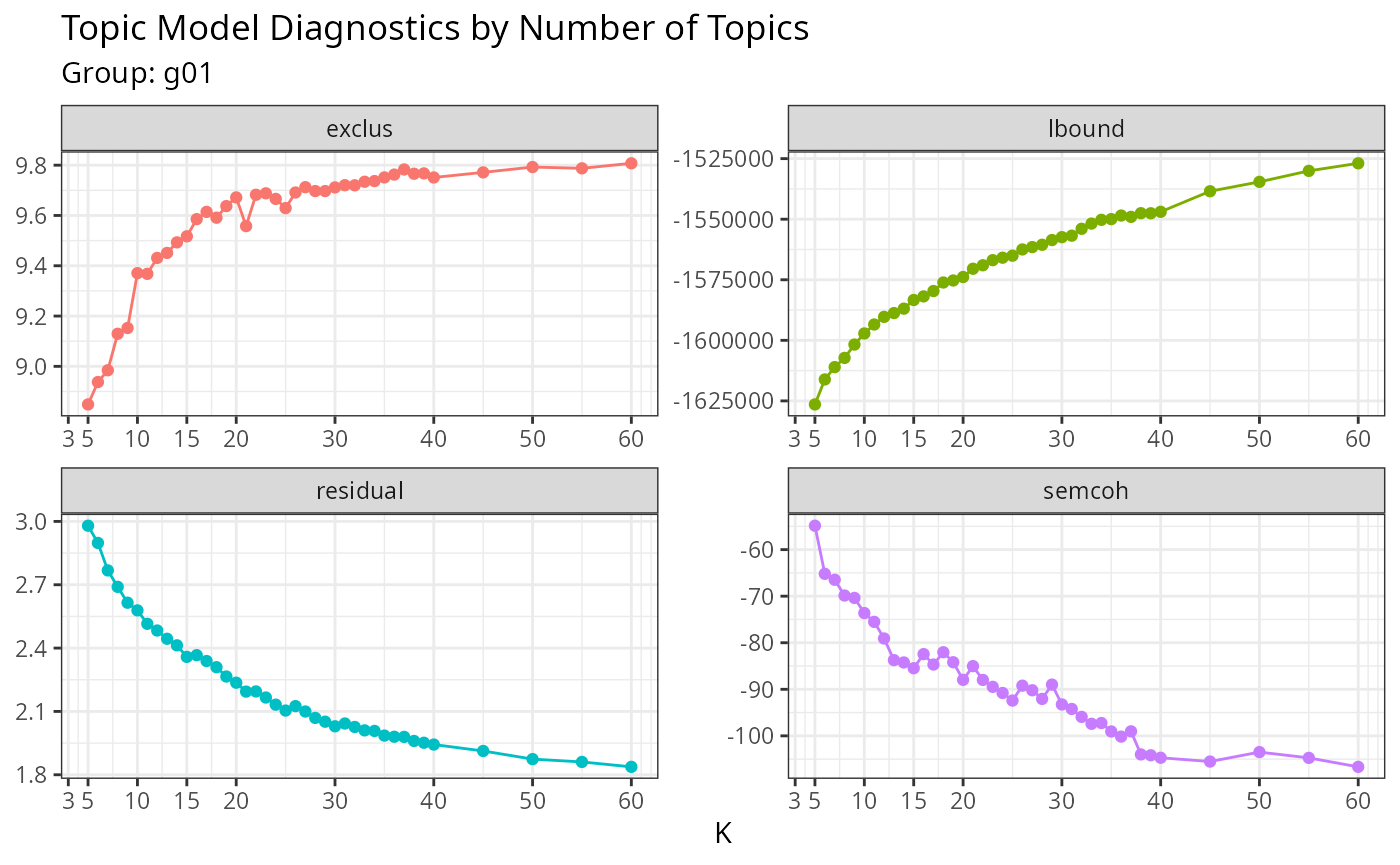
#>
#> $exclusivity_vs_coherence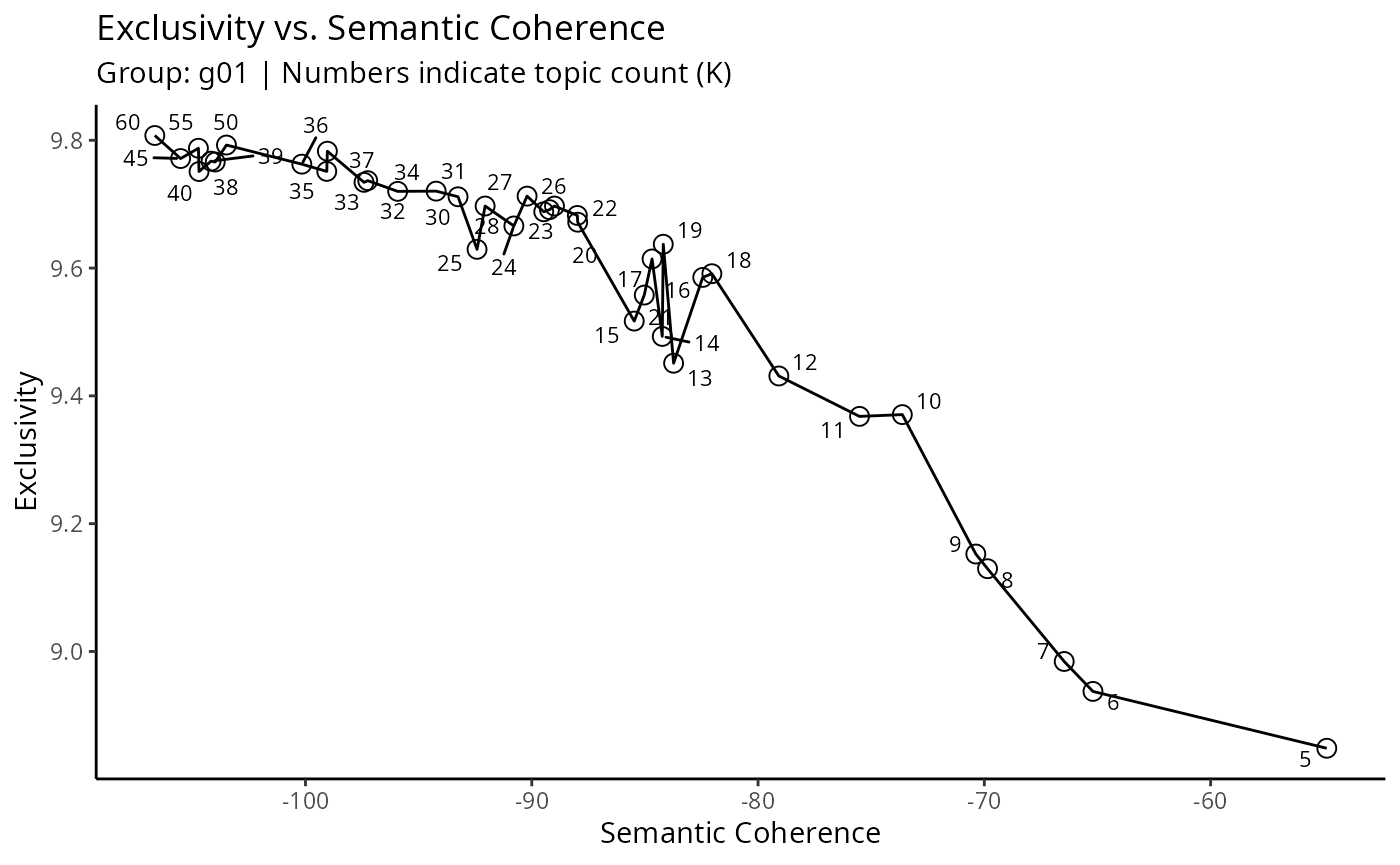
# tictoc::tic()
# groups_stm_run <- sniff_groups_stm_run(groups_stm_prepare, k_topics = 18, n_top_documents = 20)
# tictoc::toc()
# 35 sec
groups_stm_run <- readRDS('~/Sync/birddog-data/wos-sugarcane-groups-stm-run-g01.rds')
groups_stm_run$topic_proportion |>
dplyr::mutate(topic_proportion = round(topic_proportion, 3)) |>
DT::datatable(
caption = 'g01',
rownames = FALSE,
filter = 'bottom',
extensions = 'Buttons',
escape = FALSE,
options = list(dom = 'Blfrtip', pageLength = 10)
)
groups_stm_run$top_documents |>
dplyr::left_join(M |> dplyr::select(document = DI2, SR), by = dplyr::join_by(document)) |>
dplyr::mutate(SR = paste0('<a href="https://www.webofscience.com/wos/alldb/full-record/', SR, '">', SR, '</a>')) |>
dplyr::select(SR, topic, gamma, title) |>
DT::datatable(
caption = 'g01',
rownames = FALSE,
filter = 'bottom',
extensions = 'Buttons',
escape = FALSE,
options = list(dom = 'Blfrtip', pageLength = 10)
)Indexes: Citatations Cycle Time
All network.
# tictoc::tic()
# net_cct <- sniff_citations_cycle_time(net, scope = 'network', start_year = 1990, end_year = 2024)
# tictoc::toc()
# 1.3 min
net_cct <- readRDS('~/Sync/birddog-data/wos-sugarcane-net-cct.rds')
# names(net_cct)
net_cct$plots[['full_network']]Groups.
tictoc::tic()
groups_cct <- sniff_citations_cycle_time(groups, scope = 'groups', start_year = 1990, end_year = 2024)
tictoc::toc()
# 1.4 minIndexes: Entropy
All network.
tictoc::tic()
net_entropy <- sniff_entropy(net, scope = 'network', start_year = 1990, end_year = 2024)
tictoc::toc()
# 5.7 secGroups.
tictoc::tic()
groups_entropy <- sniff_entropy(groups, scope = 'groups', start_year = 1990, end_year = 2024)
tictoc::toc()
#> 8.257 sec elapsed
# 7.7 sec
groups_entropy$plots[['g02']]Main Path Analysis
Key Route Algorithm.
All network.
net_dc <- sniff_network(M, type = "direct citation")
igraph::V(net_dc)$deg <- igraph::degree(net_dc)
net_dc |>
tidygraph::activate(nodes) |>
dplyr::filter(deg >= 50) ->
net_dc_topn
net_key_route <- sniff_key_route(net_dc_topn, scope = 'network')
net_key_route[['full_network']]$plot
net_key_route[['full_network']]$data |>
dplyr::select(- name) |>
DT::datatable(
rownames = FALSE,
filter = 'bottom',
extensions = 'Buttons',
escape = FALSE,
options = list(dom = 'Blfrtip', pageLength = 5)
)Groups.
comps_dc <- birddog::sniff_components(net_dc)
groups_dc <- birddog::sniff_groups(comps_dc, algorithm = 'fast_greedy', min_group_size = 30, groups_short_name = TRUE)
groups_key_route <- sniff_key_route(groups_dc, scope = 'groups')Session info
sessioninfo::session_info()$platform |>
unlist() |>
as.data.frame() |>
tibble::rownames_to_column() |>
setNames(c("Setting", "Value")) |>
gt::gt()| Setting | Value |
|---|---|
| version | R version 4.4.3 (2025-02-28) |
| os | Manjaro Linux |
| system | x86_64, linux-gnu |
| ui | X11 |
| language | en |
| collate | en_US.UTF-8 |
| ctype | en_US.UTF-8 |
| tz | America/Cuiaba |
| date | 2025-09-16 |
| pandoc | 3.1.12.1 @ /usr/bin/ (via rmarkdown) |
| quarto | 1.4.553 @ /usr/bin/quarto |
